Difference between revisions of "JupyterHub"
Torradeflot (talk | contribs) |
|||
| (75 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | tldr; Connect to https://jupyter.pic.es/ . Enjoy! | |
= Introduction = | = Introduction = | ||
| Line 11: | Line 11: | ||
In practice that means that you should estimate the test data volume that you work with during a session to be able to be processed in less than 48 hours. | In practice that means that you should estimate the test data volume that you work with during a session to be able to be processed in less than 48 hours. | ||
| − | = How to connect to the service = | + | == How to connect to the service == |
Got to [https://jupyter.pic.es jupyter.pic.es] to see your login screen. | Got to [https://jupyter.pic.es jupyter.pic.es] to see your login screen. | ||
| − | [[File:login.png | + | [[File:login.png|700px|Login screen]] |
Sign in with your PIC user credentials. This will prompt you to the following screen. | Sign in with your PIC user credentials. This will prompt you to the following screen. | ||
| − | [[File: | + | [[File:ScreenshotJupyterSpawn.png|700px|current]] |
Here you can choose the hardware configuration for your Jupyter session. Also, you have to choose the experiment (project) you are working on during the Jupyter session. After choosing a configuration and pressing start the next screen will show you the progress of the initialisation process. Keep in mind that a job containing your Jupyter session is actually sent to the HTCondor queuing system and waiting for available resources before being started. This usually takes less than a minute but can take up to a few depending on our resource usage. | Here you can choose the hardware configuration for your Jupyter session. Also, you have to choose the experiment (project) you are working on during the Jupyter session. After choosing a configuration and pressing start the next screen will show you the progress of the initialisation process. Keep in mind that a job containing your Jupyter session is actually sent to the HTCondor queuing system and waiting for available resources before being started. This usually takes less than a minute but can take up to a few depending on our resource usage. | ||
| − | [[File:screen02.png| | + | [[File:screen02.png|900px]] |
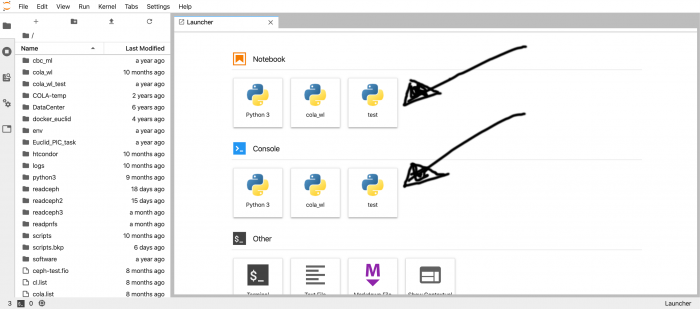
In the next screen you can choose the tool that you want to use for your work: a Python notebook, a Python console or a plain bash terminal. | In the next screen you can choose the tool that you want to use for your work: a Python notebook, a Python console or a plain bash terminal. | ||
| Line 30: | Line 30: | ||
* the XPython version of Python 3.9, this one allows you to use the integrated debugging module. | * the XPython version of Python 3.9, this one allows you to use the integrated debugging module. | ||
| − | + | == Terminate your session and logout == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | = Terminate your session and logout = | ||
It is important that you terminate your session before you log out. In order to do so, go to the top page menu "'''File''' -> '''Hub Control Panel'''" and you will see the following screen. | It is important that you terminate your session before you log out. In order to do so, go to the top page menu "'''File''' -> '''Hub Control Panel'''" and you will see the following screen. | ||
| Line 50: | Line 42: | ||
This section covers the use of Python virtual environments with Jupyter. | This section covers the use of Python virtual environments with Jupyter. | ||
| − | == | + | == Prebuilt environments == |
| + | |||
| + | PIC's jupyterhub service comes with a collection of prebuilt environments located at '''/data/jupyter/software/envs'''. | ||
| + | |||
| + | The master environment located at '''/data/jupyter/software/envs/master''' is the one used to start the jupyterlab service and the default for new notebooks. | ||
| − | + | This is a non-extensive list of the packages included: | |
| + | - astropy=6.1.0 | ||
| + | - bokeh=3.4.1 | ||
| + | - dash=2.17.0 | ||
| + | - dask=2024.5.1 | ||
| + | - findspark=2.0.1 | ||
| + | - matplotlib=3.8.4 | ||
| + | - numpy=1.26.4 | ||
| + | - pandas=2.2.2 | ||
| + | - pillow=10.3.0 | ||
| + | - plotly=5.22.0 | ||
| + | - pyhive=0.7.0 | ||
| + | - python=3.12 | ||
| + | - pywavelets=1.4.1 | ||
| + | - scikit-image=0.22.0 | ||
| + | - scikit-learn=1.5.0 | ||
| + | - scipy=1.13.1 | ||
| + | - seaborn=0.13.2 | ||
| + | - statsmodels=0.14.2 | ||
| − | + | == Initialize conda (we highly recommend the use of mamba/micromamba) == | |
| − | + | Before using conda/mamba in your bash session, you have to initialize it. | |
| + | * For access to an available conda/mamba installation, please get in contact with your project liaison at PIC. He/she will give you the actual value for the '''/path/to/conda/mamba''' placeholder. | ||
| + | * If you want to use your own conda/mamba/micromamba installation, there are two recommended options | ||
| + | ** '''miniforge''': a distribution with conda and mamba executables in a minimal base environment, instructions [https://github.com/conda-forge/miniforge here] | ||
| + | ** '''micromamba''': a self-contained executable (micromamba) with no base environment, instructions [https://mamba.readthedocs.io/en/latest/installation/micromamba-installation.html here] | ||
| + | |||
| + | Log onto Jupyter and start a session. On the homepage of your Jupyter session, click on the terminal button on the session dashboard on the right to open a bash terminal. | ||
| + | |||
| + | In order to use conda/mamba/micromamba you need to intialize the shell. This initialization can be persistent, which will do some changes to your '''~/.bashrc''' file, or you can do it every time you want to use it. | ||
| + | |||
| + | Run the following command: | ||
| + | |||
| + | <pre> | ||
| + | eval "$(/data/astro/software/miniforge3/bin/conda shell.bash hook)" | ||
| + | </pre> | ||
| + | |||
| + | Then, if you are using miniforge and you want to persist the initialization: | ||
<pre> | <pre> | ||
| − | + | /data/astro/software/miniforge3/bin/mamba init | |
</pre> | </pre> | ||
| Line 65: | Line 95: | ||
<pre> | <pre> | ||
[neissner@td110 ~]$ conda config --set auto_activate_base false | [neissner@td110 ~]$ conda config --set auto_activate_base false | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</pre> | </pre> | ||
| Line 175: | Line 200: | ||
You can use pip install inside a mamba environment, however, resolving dependencies might require installing additional packages manually. | You can use pip install inside a mamba environment, however, resolving dependencies might require installing additional packages manually. | ||
| − | = | + | == Conda / mamba configuration == |
| − | + | The behaviour of conda/mamba can be configured through the "$HOME/.condarc" file, described [https://docs.conda.io/projects/conda/en/latest/configuration.html here]. Some interesting parameters: | |
| + | |||
| + | * envs_dirs: The list of directories to search for named environments. E.g.: different locations where you created environments | ||
| + | |||
| + | envs_dirs: | ||
| + | - /data/pic/scratch/torradeflot/envs | ||
| + | - /data/astro/scratch/torradeflot/envs | ||
| + | - /data/aai/scratch/torradeflot/envs | ||
| + | |||
| + | * pkgs_dirs: Folder where to store conda packages | ||
| + | |||
| + | pkgs_dirs: | ||
| + | - /data/aai/scratch_ssd/torradeflot/pkgs | ||
| + | - /data/aai/scratch/torradeflot/pkgs | ||
| + | - /data/pic/scratch/torradeflot/pkgs | ||
| + | |||
| + | if `pkgs_dirs` and `envs_dirs` are in the same storage, conda will use hard links, thus optimizing the disk space. | ||
| + | |||
| + | |||
| + | = Jupyterlab user guide = | ||
| + | |||
| + | You can find the official documentation of the currently installed version of jupyterlab (3.6) [https://jupyterlab.readthedocs.io/en/4.2.x/ here], there you will find instruction on how to | ||
| + | * [https://jupyterlab.readthedocs.io/en/4.2.x/user/commands.html Access the command palette] | ||
| + | * [https://jupyterlab.readthedocs.io/en/4.2.x/user/toc.html Build a Table Of Contents] | ||
| + | * [https://jupyterlab.readthedocs.io/en/4.2.x/user/debugger.html Debug your code] | ||
| + | |||
| + | A set of non-official jupyterlab extensions are installed to provide additional functionalities | ||
| + | |||
| + | == Sidecar Apps: Remote desktop & Visual Studio IDE == | ||
| + | Further you see an icon with a "D" - desktop, this one starts a VNC session that allows the use of programs with graphical user interfaces. | ||
| + | |||
| + | You can also find the icon of Visual Studio, an integrated development environment. | ||
| + | |||
| + | [[File:ScreenshotJupyterlab20231103.png|700px]] | ||
| + | |||
| + | == Jupytext == | ||
| + | Pair your notebooks with text files to enhance version tracking. | ||
| + | https://jupytext.readthedocs.io | ||
| + | |||
| + | '''Example''' | ||
| − | + | If you had a notebook (.ipynb file) containing only the cell below, tracked in a git repository | |
<pre> | <pre> | ||
| − | + | %matplotlib inline | |
| − | + | import numpy as np | |
| − | [ | + | import matplotlib.pyplot as plt |
| − | + | plt.imshow(np.random.random([10, 10])) | |
</pre> | </pre> | ||
| − | + | Different executions of the cell would produce different images, and the images are embedded in a pseudo-binary format inside the notebook file. In this case, doing a '''git diff''' of the .ipynb file would produce a huge output (because the image changed), even if there wasn't any change in the code. It is thus convenient to sync the notebook with a text file (e.g. a .py script) using the jupytext extension and track this one with git. The outputs, including images, as well as some additional metadata, won't be added to the synced text file. So in the case of different executions of the same notebook, the diff will always be empty. | |
| + | |||
| + | == Git == | ||
| + | Sidebar GUI to git repo management | ||
| + | https://github.com/jupyterlab/jupyterlab-git | ||
| + | |||
| + | == Variable inspector == | ||
| + | Variable Inspector provides an interactive interface for inspecting the current state of variables in a JupyterLab session. It allows users to view variable names, types, shapes, and values in a structured table, facilitating exploratory analysis and debugging workflows similar to variable inspection tools available in environments such as MATLAB. | ||
| + | |||
| + | https://github.com/jupyterlab-contrib/jupyterlab-variableInspector | ||
| + | |||
| + | == Jupyter server proxy == | ||
| + | |||
| + | This extension is installed in PIC's jupyter environment and it is used to be able to access network/web services running on the same host as the jupyterlab server from outside through the "https://jupyter.pic.es/user/{username}/proxy/{port}" URL. | ||
| + | |||
| + | Full documentation here: https://jupyter-server-proxy.readthedocs.io/en/latest/index.html | ||
| + | ... | ||
| + | |||
| + | |||
| + | = Tips & Tricks = | ||
| + | |||
| + | == Software of particular interest == | ||
| + | |||
| + | === ROOT === | ||
| + | |||
| + | Using ROOT from a jupyter notebook needs to do some tricks. | ||
| + | |||
| + | Environment creation and kernel installation | ||
| + | |||
| + | $ micromamba env create -p /data/pic/scratch/torradeflot/envs/mcdata root ipykernel | ||
| + | $ micromamba activate mcdata | ||
| + | $ python -m ipykernel install --user --name mcdata | ||
| + | |||
| + | After doing this ROOT can be imported from a python shell, but it does not work from a notebook. | ||
| + | |||
| + | * ROOT uses JIT compilation with Cling: https://root.cern/cling/ | ||
| + | * conda provides it's own set of compilation tools: gcc, gxx, fortran | ||
| + | * Some environment variables are necessary to ensure that these two pieces work well together: | ||
| + | ** PATH: to be able to find the compiler tools | ||
| + | ** CONDA_BUILD_SYSROOT: needed to configure the compiler call | ||
| + | |||
| + | These environment variables are not propagated to the notebook, because they are set by .bashrc conda activate, ... so we need to explicitly set them in the notebook. | ||
| − | + | import os | |
| − | + | import sys | |
| − | + | from pathlib import Path | |
| − | [ | + | bin_dir = Path(sys.executable).parent |
| − | [ | + | os.environ['PATH'] = f'{bin_dir}:{os.environ['PATH']}' |
| − | + | os.environ['CONDA_BUILD_SYSROOT'] = str(bin_dir.parent / 'x86_64-conda-linux-gnu/sysroot') | |
| − | |||
| − | = | + | === SageMath === |
| − | == | ||
[https://www.sagemath.org/ SageMath] is particularly interesting for Cosmology because it allows symbolic calculations, e.g. deriving the equations of motions for the scale factor starting from a customised space-time metric. | [https://www.sagemath.org/ SageMath] is particularly interesting for Cosmology because it allows symbolic calculations, e.g. deriving the equations of motions for the scale factor starting from a customised space-time metric. | ||
| − | + | '''Standard cosmology examples''' | |
* The Friedman equations for the FLRW solution of the Einstein equations. | * The Friedman equations for the FLRW solution of the Einstein equations. | ||
| Line 216: | Line 319: | ||
[[File:Screenshot_Sage06.png|300px]] | [[File:Screenshot_Sage06.png|300px]] | ||
| − | + | '''Enabling SageMath environment in Jupyter''' | |
If you have never initialized mamba, run: | If you have never initialized mamba, run: | ||
| Line 256: | Line 359: | ||
Next time you go to your Jupyter dashboard you will find the sage environment listed there. | Next time you go to your Jupyter dashboard you will find the sage environment listed there. | ||
| + | |||
| + | == Dask == | ||
| + | Dask supports parallel computations in Python. The PIC Jupyterlab has an extension for launching | ||
| + | your own Dask clusters. For more information, see [[Dask|Dask documentation]]. | ||
| + | |||
| + | |||
| + | == Using a Singularity image as a Jupyter kernel == | ||
| + | |||
| + | In some projects, the software stack is provided as a Singularity image. In such cases, it can be convenient to use this image directly as a Jupyter kernel, allowing notebooks on jupyter.pic.es to run within the same controlled software environment. | ||
| + | |||
| + | To be used as a Jupyter kernel, the Singularity image must satisfy certain requirements. These depend on the programming language used inside the notebook. | ||
| + | |||
| + | The singularity image needs to have the '''python''' and the '''ipykernel''' module installed. | ||
| + | |||
| + | * Create the folder that will host the kernel definition | ||
| + | |||
| + | mkdir -p $HOME/.local/share/jupyter/kernels/singularity | ||
| + | |||
| + | * Create the '''kernel.json''' file inside it with the following content: | ||
| + | |||
| + | { | ||
| + | "argv": [ | ||
| + | "singularity", | ||
| + | "exec", | ||
| + | "--cleanenv", | ||
| + | "/path/to/the/singularity/image.sif", | ||
| + | "python", | ||
| + | "-m", | ||
| + | "ipykernel", | ||
| + | "-f", | ||
| + | "{connection_file}" | ||
| + | ], | ||
| + | "language": "python", | ||
| + | "display_name": "singularity-kernel" | ||
| + | } | ||
| + | |||
| + | Refresh or start the jupyterlab interface and the singularity kernel should appear in the launcher tab | ||
| + | |||
| + | == GPUs == | ||
| + | |||
| + | The way to identify the GPUs that are assigned to your job is: | ||
| + | * check the environment variable CUDA_VISIBLE_DEVICES. In a terminal run "echo $CUDA_VISIBLE_DEVICES". The environment variable contains a list of comma-separated GPU ids. With this you will already know how many gpus are assigned to your job. If the variable does not exist, there are no gpus assigned to the job | ||
| + | |||
| + | * list the gpus with nvidia-smi, in a terminal run "nvidia-smi -L", and look for the gpus you've been assigned. Remember their indexes (integers from 0 to 7) | ||
| + | |||
| + | * The two steps above can be done with the following command: | ||
| + | |||
| + | nvidia-smi -L | grep $CUDA_VISIBLE_DEVICES | ||
| + | |||
| + | if only having a single assigned GPU. | ||
| + | |||
| + | [[File:check_gpu_id_highlighted.png]] | ||
| + | |||
| + | * in the GPU dashboard the gpus are identified with their index | ||
| + | |||
| + | [[File:check_gpu_resources_highlighted.png]] | ||
| + | |||
| + | |||
| + | == Code samples == | ||
| + | |||
| + | A repository with sample code can be found here: https://gitlab.pic.es/services/code-samples/ | ||
| + | |||
| + | == Running notebooks through HTCondor == | ||
| + | After developing a notebook, you might want to run it as a script with different configurations. The | ||
| + | following documentation explains | ||
| + | |||
| + | [[notebook_htcondor|how to run a notebook through HTCondor.]] | ||
| + | |||
| + | = Troubleshooting = | ||
| + | |||
| + | == Logs == | ||
| + | |||
| + | The log files for the jupyterlab server are stored in "~/.jupyter". The log files will be created once the jupyter lab server job is finished. | ||
| + | |||
| + | |||
| + | == Clean workspaces == | ||
| + | |||
| + | Jupyterlab stores the workspace status in the "~/.jupyter/lab/workspaces" folder. If you want to start with a fresh (empty) workspace, delete all the content of this folder before launching the notebook. | ||
| + | |||
| + | cd ~/.jupyter/lab/workspaces | ||
| + | rm * | ||
| + | |||
| + | = Known problems = | ||
| + | |||
| + | == Error 500: Internal Server Error == | ||
| + | |||
| + | This is a generic error. Means that the jupyterlab server failed. This could be for different reasons: | ||
| + | |||
| + | * Your HOME folder is full. Log in to "ui.pic.es" and run "quota" to check the usage vs quota. If it is full you'll have to free up space. | ||
| + | |||
| + | == 504 Gateway timeout == | ||
| + | |||
| + | The notebook job is running in HTCondor but the user can not access the notebook server. Ultimately a 504 error is received. | ||
| + | |||
| + | This is probably because there's some error when starting the jupyterlab server. First of all, shutdown the notebook server and [[#Logs|check the logs]] to better identify the problem. If you don't see the source of the error, try to [[#Clean_workspaces|clean the workspaces]] and launch a notebook again. | ||
| + | |||
| + | == Loading libraries is very slow == | ||
| + | Conda environments at storage using hard drives can be extremely slow to load. If you encounter this problem, please ask | ||
| + | your support contact for a "SSD scratch" location to store environments. Currently this service is being tested and | ||
| + | deployed as needed. | ||
| + | |||
| + | == Spawn failed: The 'ip' trait of a PICCondorSpawner instance expected a unicode string, not the NoneType None == | ||
| + | |||
| + | Jupyterhub could not get the host name from HTCondor's stdout, because it didn't match the expected regular expression. | ||
| + | |||
| + | This error happens randomly from time to time, it does not imply any major problem in any of the services. | ||
| + | |||
| + | Try to request a new notebook server. | ||
| + | |||
| + | == 403 : Forbidden. XSRF cookie does not match POST argument == | ||
| + | |||
| + | The value of the "_xsrf" cookie sent by the browser does not match the expected value. This could be for many reasons: temporary high load on the server, race conditions, temporary network unstability, many open tabs in the browser, etc. | ||
| + | |||
| + | In general it can be solved by closing all tabs pointing to "jupyter.pic.es", cleaning the cookies and connecting back to "jupyter.pic.es" | ||
| + | |||
| + | == Proper usage of X509 based proxies == | ||
| + | |||
| + | We found recently that the usage of proxies within a Jupyter session might cause problems because the environment changes certain standard locations such as '''/tmp''' | ||
| + | |||
| + | For a correct functioning please create the proxy the following way, example for Virgo: | ||
| + | |||
| + | <pre> | ||
| + | [<user>@<hostname> ~]$ /bin/voms-proxy-init --voms virgo:/virgo/ligo --out ./x509up_u$(id -u) | ||
| + | [<user>@<hostname> ~]$ export X509_USER_PROXY=./x509up_u$(id -u) | ||
| + | [<user>@<hostname> ~]$ ls /cvmfs/ligo.osgstorage.org | ||
| + | ls: cannot access /cvmfs/ligo.osgstorage.org: Permission denied | ||
| + | </pre> | ||
| + | |||
| + | Here the proxy cannot properly be located. Therefore we have to put the complete path into the variable: | ||
| + | |||
| + | <pre> | ||
| + | [<user>@<hostname> ~]$ pwd | ||
| + | /nfs/pic.es/user/<letter>/<user> | ||
| + | [<user>@<hostname> ~]$ export X509_USER_PROXY=/nfs/pic.es/user/<letter>/<user>/x509up_u$(id -u) | ||
| + | [<user>@<hostname> ~]$ ls /cvmfs/ligo.osgstorage.org | ||
| + | frames powerflux pycbc test_access | ||
| + | </pre> | ||
Latest revision as of 06:21, 15 January 2026
tldr; Connect to https://jupyter.pic.es/ . Enjoy!
Introduction
PIC offers a service for running Jupyter notebooks on CPU or GPU resources. This service is primarily thought for code developing or prototyping rather than data processing. The usage is similar to running notebooks on your personal computer but offers the advantage of developing and testing your code on different hardware configurations, as well as facilitating the scalability of the code since it is being tested in the same environment in which it would run on a mass scale.
Since the service is strictly thought for development and small scale testing tasks, a shutdown policy for the sessions has been put in place:
- The maximum duration for a session is 48h.
- After an idle period of 2 hours, the session will be closed.
In practice that means that you should estimate the test data volume that you work with during a session to be able to be processed in less than 48 hours.
How to connect to the service
Got to jupyter.pic.es to see your login screen.
Sign in with your PIC user credentials. This will prompt you to the following screen.
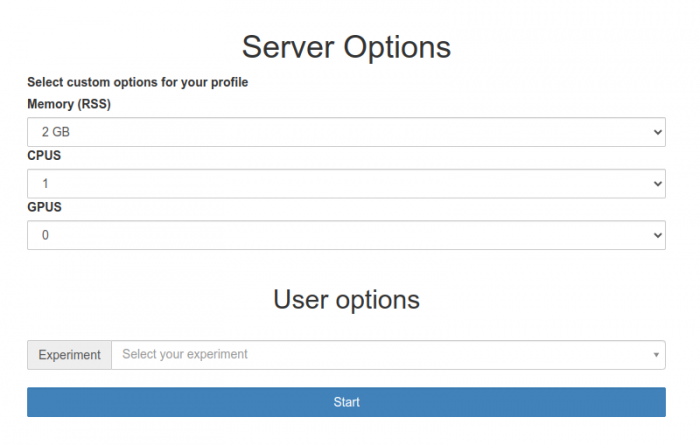
Here you can choose the hardware configuration for your Jupyter session. Also, you have to choose the experiment (project) you are working on during the Jupyter session. After choosing a configuration and pressing start the next screen will show you the progress of the initialisation process. Keep in mind that a job containing your Jupyter session is actually sent to the HTCondor queuing system and waiting for available resources before being started. This usually takes less than a minute but can take up to a few depending on our resource usage.
In the next screen you can choose the tool that you want to use for your work: a Python notebook, a Python console or a plain bash terminal. For the Python environment (either notebook or environment) you have two default options:
- the ipykernel version of Python 3
- the XPython version of Python 3.9, this one allows you to use the integrated debugging module.
Terminate your session and logout
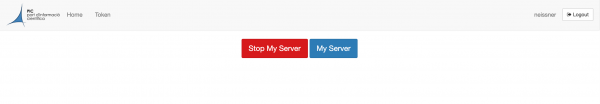
It is important that you terminate your session before you log out. In order to do so, go to the top page menu "File -> Hub Control Panel" and you will see the following screen.
Here, click on the Stop My Server button. After that you can log out by clicking the Logout button in the right upper corner.
Python virtual environments
This section covers the use of Python virtual environments with Jupyter.
Prebuilt environments
PIC's jupyterhub service comes with a collection of prebuilt environments located at /data/jupyter/software/envs.
The master environment located at /data/jupyter/software/envs/master is the one used to start the jupyterlab service and the default for new notebooks.
This is a non-extensive list of the packages included:
- astropy=6.1.0 - bokeh=3.4.1 - dash=2.17.0 - dask=2024.5.1 - findspark=2.0.1 - matplotlib=3.8.4 - numpy=1.26.4 - pandas=2.2.2 - pillow=10.3.0 - plotly=5.22.0 - pyhive=0.7.0 - python=3.12 - pywavelets=1.4.1 - scikit-image=0.22.0 - scikit-learn=1.5.0 - scipy=1.13.1 - seaborn=0.13.2 - statsmodels=0.14.2
Initialize conda (we highly recommend the use of mamba/micromamba)
Before using conda/mamba in your bash session, you have to initialize it.
- For access to an available conda/mamba installation, please get in contact with your project liaison at PIC. He/she will give you the actual value for the /path/to/conda/mamba placeholder.
- If you want to use your own conda/mamba/micromamba installation, there are two recommended options
Log onto Jupyter and start a session. On the homepage of your Jupyter session, click on the terminal button on the session dashboard on the right to open a bash terminal.
In order to use conda/mamba/micromamba you need to intialize the shell. This initialization can be persistent, which will do some changes to your ~/.bashrc file, or you can do it every time you want to use it.
Run the following command:
eval "$(/data/astro/software/miniforge3/bin/conda shell.bash hook)"
Then, if you are using miniforge and you want to persist the initialization:
/data/astro/software/miniforge3/bin/mamba init
This actually changes the .bashrc file in your home directory in order to activate the base environment on login. To avoid that the base environment is activated every time you log on to a node, run:
[neissner@td110 ~]$ conda config --set auto_activate_base false
Link an existing environment to Jupyter
You can find instructions on how to create your own environments, e.g. here.
Log into Jupyter, start a session. From the session dashboard choose the bash terminal.
Inside the terminal, activate your environment.
For conda/mamba environments:
- if you created the environment without a prefix:
[neissner@td110 ~]$ mamba activate environment (...) [neissner@td110 ~]$
The parenthesis (...) in front of your bash prompt show the name of your environment.
- if you created the environment with a prefix:
[neissner@td110 ~]$ mamba activate /path/to/environment (...) [neissner@td110 ~]$
The parenthesis (...) in front of your bash prompt show the absolute path of your environment.
For venv environments:
[neissner@td110 ~]$ source /path/to/environment/bin/activate (...) [neissner@td110 ~]$
Link the environment to a Jupyter kernel. For both, conda/mamba and venv:
(...) [neissner@td110 ~]$ python -m ipykernel install --user --name=whatever_kernel_name
Installed kernelspec whatever_kernel_name in
/nfs/pic.es/user/n/neissner/.local/share/jupyter/kernels/whatever_kernel_name
If you don't have the ipykernel module installed in your environment you may receive an error message like the one below when trying to run the previous command.
No module named ipykernel
If this is the case, you need to install it by running: pip install ipykernel
Deactivate your environment.
For conda:
(...) [neissner@td110 ~]$ mamba deactivate
For venv:
(...) [neissner@td110 ~]$ deactivate
Now you can exit the terminal. After refreshing the Jupyter page your whatever_kernel_name appears in the dashboard. In this example test has been used for whatever_kernel_name
Unlink an environment from Jupyter
Log onto Jupyter, start a session and from the session dashboard choose the bash terminal. To remove your environment/kernel from Jupyter run:
[neissner@td110 ~]$ jupyter kernelspec uninstall whatever_kernel_name Kernel specs to remove: whatever_kernel_name /nfs/pic.es/user/n/neissner/.local/share/jupyter/kernels/whatever_kernel_name Remove 1 kernel specs [y/N]: y [RemoveKernelSpec] Removed /nfs/pic.es/user/n/neissner/.local/share/jupyter/kernels/whatever_kernel_name
Keep in mind that, although not available in Jupyter anymore, the environment still exists. Whenever you need it, you can link it again.
Create virtual environments with venv or conda
Before creating a new environment, please get in contact with your project liaison at PIC as there may be already a suitable environment for your needs in place.
If none of the existing environments suits your needs, you can create a new environment. First, create a directory in a suitable place to store the environment. For single-user environments, place them in your home under ~/env. For environments that will be shared with other project users, contact your project liaison and ask him/her for a path in a shared storage volume that is visible to all of them.
Once you have the location (i.e. /path/to/env/folder), create the environment with the following commands:
For venv environments (recommended)
If your_env is installed at /path/to/env/your_env
[neissner@td110 ~]$ cd /path/to/env [neissner@td110 ~]$ python3 -m venv your_env
Now you should be able to activate your environment and install additional modules
[neissner@td110 ~]$ cd /path/to/env [neissner@td110 ~]$ source your_env/bin/activate (...)[neissner@td110 ~]$ pip install additional_module1 additional_module2 ...
For conda/mamba environments
[neissner@td110 ~]$ mamba create --prefix /path/to/env/your_env
The list of modules (module1, module2, ...) is optional. For instance, for a python3 environment with scipy you would specify: python=3 scipy
Now you should be able to activate your environment and install additional modules
[neissner@td110 ~]$ mamba activate /path/to/env/folder/your_env (...)[neissner@td110 ~]$ mamba install additional_module1 additional_module2 ...
You can use pip install inside a mamba environment, however, resolving dependencies might require installing additional packages manually.
Conda / mamba configuration
The behaviour of conda/mamba can be configured through the "$HOME/.condarc" file, described here. Some interesting parameters:
- envs_dirs: The list of directories to search for named environments. E.g.: different locations where you created environments
envs_dirs: - /data/pic/scratch/torradeflot/envs - /data/astro/scratch/torradeflot/envs - /data/aai/scratch/torradeflot/envs
- pkgs_dirs: Folder where to store conda packages
pkgs_dirs: - /data/aai/scratch_ssd/torradeflot/pkgs - /data/aai/scratch/torradeflot/pkgs - /data/pic/scratch/torradeflot/pkgs
if `pkgs_dirs` and `envs_dirs` are in the same storage, conda will use hard links, thus optimizing the disk space.
Jupyterlab user guide
You can find the official documentation of the currently installed version of jupyterlab (3.6) here, there you will find instruction on how to
A set of non-official jupyterlab extensions are installed to provide additional functionalities
Sidecar Apps: Remote desktop & Visual Studio IDE
Further you see an icon with a "D" - desktop, this one starts a VNC session that allows the use of programs with graphical user interfaces.
You can also find the icon of Visual Studio, an integrated development environment.
Jupytext
Pair your notebooks with text files to enhance version tracking. https://jupytext.readthedocs.io
Example
If you had a notebook (.ipynb file) containing only the cell below, tracked in a git repository
%matplotlib inline import numpy as np import matplotlib.pyplot as plt plt.imshow(np.random.random([10, 10]))
Different executions of the cell would produce different images, and the images are embedded in a pseudo-binary format inside the notebook file. In this case, doing a git diff of the .ipynb file would produce a huge output (because the image changed), even if there wasn't any change in the code. It is thus convenient to sync the notebook with a text file (e.g. a .py script) using the jupytext extension and track this one with git. The outputs, including images, as well as some additional metadata, won't be added to the synced text file. So in the case of different executions of the same notebook, the diff will always be empty.
Git
Sidebar GUI to git repo management https://github.com/jupyterlab/jupyterlab-git
Variable inspector
Variable Inspector provides an interactive interface for inspecting the current state of variables in a JupyterLab session. It allows users to view variable names, types, shapes, and values in a structured table, facilitating exploratory analysis and debugging workflows similar to variable inspection tools available in environments such as MATLAB.
https://github.com/jupyterlab-contrib/jupyterlab-variableInspector
Jupyter server proxy
This extension is installed in PIC's jupyter environment and it is used to be able to access network/web services running on the same host as the jupyterlab server from outside through the "https://jupyter.pic.es/user/{username}/proxy/{port}" URL.
Full documentation here: https://jupyter-server-proxy.readthedocs.io/en/latest/index.html ...
Tips & Tricks
Software of particular interest
ROOT
Using ROOT from a jupyter notebook needs to do some tricks.
Environment creation and kernel installation
$ micromamba env create -p /data/pic/scratch/torradeflot/envs/mcdata root ipykernel $ micromamba activate mcdata $ python -m ipykernel install --user --name mcdata
After doing this ROOT can be imported from a python shell, but it does not work from a notebook.
- ROOT uses JIT compilation with Cling: https://root.cern/cling/
- conda provides it's own set of compilation tools: gcc, gxx, fortran
- Some environment variables are necessary to ensure that these two pieces work well together:
- PATH: to be able to find the compiler tools
- CONDA_BUILD_SYSROOT: needed to configure the compiler call
These environment variables are not propagated to the notebook, because they are set by .bashrc conda activate, ... so we need to explicitly set them in the notebook.
import os
import sys
from pathlib import Path
bin_dir = Path(sys.executable).parent
os.environ['PATH'] = f'{bin_dir}:{os.environ['PATH']}'
os.environ['CONDA_BUILD_SYSROOT'] = str(bin_dir.parent / 'x86_64-conda-linux-gnu/sysroot')
SageMath
SageMath is particularly interesting for Cosmology because it allows symbolic calculations, e.g. deriving the equations of motions for the scale factor starting from a customised space-time metric.
Standard cosmology examples
- The Friedman equations for the FLRW solution of the Einstein equations.
You can find the corresponding Notebook in any PIC terminal at /data/astro/software/notebooks/FLRW_cosmology.ipynb
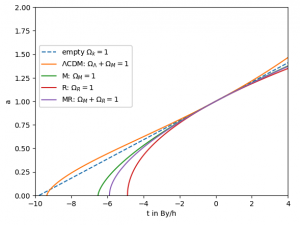
- The notebook you can find at /data/astro/software/notebooks/FLRW_cosmology_solutions.ipynb uses known analytical solutions of the FLRW cosmology and produces this image for the evolution of the scale factor:
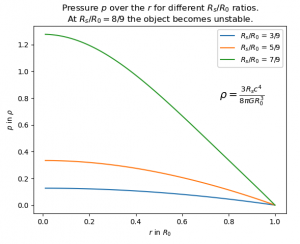
- The notebook you can find at /data/astro/software/notebooks/Interior_Schwarzschild.ipynb shows the formalism for the interior Schwarzschild metric and displays the solutions for density and pressure of a static celestial object that is sufficiently larger than its Schwarzschild radius. The pressure for an object with constant density id shown in the image:
Enabling SageMath environment in Jupyter
If you have never initialized mamba, run:
[<user>@<hostname> ~]$ /data/astro/software/centos7/conda/mambaforge_4.14.0/bin/mamba init [<user>@<hostname> ~]$ conda config --set auto_activate_base false
After that you can enable SageMath for its use in a Jupyter notebook session:
[<user>@<hostname> ~] mamba activate /data/astro/software/envs/sage (/data/astro/software/envs/sage) [<user>@<hostname> ~]$ python -m ipykernel install --user --name=sage .... (/data/astro/software/envs/sage) [<user>@<hostname> ~]$ mamba deactivate
This creates a file in you home ~/.local/share/jupyter/kernels/sage/kernel.json which has to be modified to look like this:
{
"argv": [
"/data/astro/software/envs/sage/bin/sage",
"--python",
"-m",
"sage.repl.ipython_kernel",
"-f",
"{connection_file}"
],
"display_name": "sage",
"language": "sage",
"metadata": {
"debugger": true
}
}
Next time you go to your Jupyter dashboard you will find the sage environment listed there.
Dask
Dask supports parallel computations in Python. The PIC Jupyterlab has an extension for launching your own Dask clusters. For more information, see Dask documentation.
Using a Singularity image as a Jupyter kernel
In some projects, the software stack is provided as a Singularity image. In such cases, it can be convenient to use this image directly as a Jupyter kernel, allowing notebooks on jupyter.pic.es to run within the same controlled software environment.
To be used as a Jupyter kernel, the Singularity image must satisfy certain requirements. These depend on the programming language used inside the notebook.
The singularity image needs to have the python and the ipykernel module installed.
- Create the folder that will host the kernel definition
mkdir -p $HOME/.local/share/jupyter/kernels/singularity
- Create the kernel.json file inside it with the following content:
{
"argv": [
"singularity",
"exec",
"--cleanenv",
"/path/to/the/singularity/image.sif",
"python",
"-m",
"ipykernel",
"-f",
"{connection_file}"
],
"language": "python",
"display_name": "singularity-kernel"
}
Refresh or start the jupyterlab interface and the singularity kernel should appear in the launcher tab
GPUs
The way to identify the GPUs that are assigned to your job is:
- check the environment variable CUDA_VISIBLE_DEVICES. In a terminal run "echo $CUDA_VISIBLE_DEVICES". The environment variable contains a list of comma-separated GPU ids. With this you will already know how many gpus are assigned to your job. If the variable does not exist, there are no gpus assigned to the job
- list the gpus with nvidia-smi, in a terminal run "nvidia-smi -L", and look for the gpus you've been assigned. Remember their indexes (integers from 0 to 7)
- The two steps above can be done with the following command:
nvidia-smi -L | grep $CUDA_VISIBLE_DEVICES
if only having a single assigned GPU.
- in the GPU dashboard the gpus are identified with their index
Code samples
A repository with sample code can be found here: https://gitlab.pic.es/services/code-samples/
Running notebooks through HTCondor
After developing a notebook, you might want to run it as a script with different configurations. The following documentation explains
how to run a notebook through HTCondor.
Troubleshooting
Logs
The log files for the jupyterlab server are stored in "~/.jupyter". The log files will be created once the jupyter lab server job is finished.
Clean workspaces
Jupyterlab stores the workspace status in the "~/.jupyter/lab/workspaces" folder. If you want to start with a fresh (empty) workspace, delete all the content of this folder before launching the notebook.
cd ~/.jupyter/lab/workspaces rm *
Known problems
Error 500: Internal Server Error
This is a generic error. Means that the jupyterlab server failed. This could be for different reasons:
- Your HOME folder is full. Log in to "ui.pic.es" and run "quota" to check the usage vs quota. If it is full you'll have to free up space.
504 Gateway timeout
The notebook job is running in HTCondor but the user can not access the notebook server. Ultimately a 504 error is received.
This is probably because there's some error when starting the jupyterlab server. First of all, shutdown the notebook server and check the logs to better identify the problem. If you don't see the source of the error, try to clean the workspaces and launch a notebook again.
Loading libraries is very slow
Conda environments at storage using hard drives can be extremely slow to load. If you encounter this problem, please ask your support contact for a "SSD scratch" location to store environments. Currently this service is being tested and deployed as needed.
Spawn failed: The 'ip' trait of a PICCondorSpawner instance expected a unicode string, not the NoneType None
Jupyterhub could not get the host name from HTCondor's stdout, because it didn't match the expected regular expression.
This error happens randomly from time to time, it does not imply any major problem in any of the services.
Try to request a new notebook server.
403 : Forbidden. XSRF cookie does not match POST argument
The value of the "_xsrf" cookie sent by the browser does not match the expected value. This could be for many reasons: temporary high load on the server, race conditions, temporary network unstability, many open tabs in the browser, etc.
In general it can be solved by closing all tabs pointing to "jupyter.pic.es", cleaning the cookies and connecting back to "jupyter.pic.es"
Proper usage of X509 based proxies
We found recently that the usage of proxies within a Jupyter session might cause problems because the environment changes certain standard locations such as /tmp
For a correct functioning please create the proxy the following way, example for Virgo:
[<user>@<hostname> ~]$ /bin/voms-proxy-init --voms virgo:/virgo/ligo --out ./x509up_u$(id -u) [<user>@<hostname> ~]$ export X509_USER_PROXY=./x509up_u$(id -u) [<user>@<hostname> ~]$ ls /cvmfs/ligo.osgstorage.org ls: cannot access /cvmfs/ligo.osgstorage.org: Permission denied
Here the proxy cannot properly be located. Therefore we have to put the complete path into the variable:
[<user>@<hostname> ~]$ pwd /nfs/pic.es/user/<letter>/<user> [<user>@<hostname> ~]$ export X509_USER_PROXY=/nfs/pic.es/user/<letter>/<user>/x509up_u$(id -u) [<user>@<hostname> ~]$ ls /cvmfs/ligo.osgstorage.org frames powerflux pycbc test_access